Dr. Vibin Ipe Thomas
Education
- BSc., MG University, 2003
- MSc., MG University, 2005
- Ph.D., University of Montreal, 2012
- Post-doc, Stanford University, USA, 2013-14
Area of Specialization
- Physical
- Theoretical
- Computational
Modern theoretical methods combined with advanced scientific computing have transformed our ability to perform modeling and simulation studies of key processes like proton and electron transfer reactions in chemistry, material science and biology that generate realistic results with full atomic resolution.
Research efforts in my group revolves around understanding elementary proton and electron transfer reactions through state-of-art computer simulations. Proton transfer reactions are mainly studied using state-of-art ab initio molecular dynamics simulations, including a DF-QM method (for studying the acid base reaction and to compare with the experimental photo-acid ultrasft spectroscopic investigations), ab initio-centroid molecular dynamics simulations (to include nuclear quantum effects), Ring polymer molecular dynamics simulations (to include nuclear quantum effects), hybrid QM/MM Molecular dynamics simulations. The electron transfer reactions are mainly investigated using surface hopping technique.
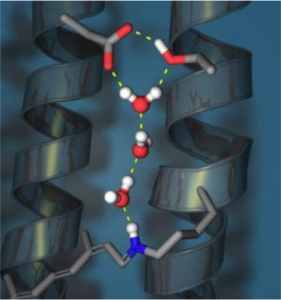
Proton Transfer investigations in Biological System.
Focus: Understanding the microscopic proton transport mechanism in biological Membranes
Proton transport in proteins through proton channels is very important for sustaining life. A crucial example is M2 proton channel of influenza A virus, which is a prototype for a class of viral ion channel known as viroporines, conduct acid gated proton along a chain of water molecule and ionisable side chains. This enables the hydrogen ions to enter the viral particles. This proton channel plays a crucial role in enabling the virus to infect and replicate in other processes as illustrated by many experimentalists. The importance of the investigation of proton channels in influenza A proton channel can be understood by the fact that a mutation in the M2 proton channel, Ser31 to Asn is responsible for the drug resistance of the recent HINI swine flu epidemic as the drug was targeting on blocking the proton transfer pathway. A detailed molecular level understanding of the proton transport in M2 proton channel will not only provide information about the Influenza virus infection process by also provide rich source of information on the proton transfer through biological membranes.
Electron Transfer Investigations
Focus: Develop and apply computational tools for studying the electron transfer dynamics in dye-sensitized solar cells
In the last 50 years bulk silicon solar cells, developed to convert sunlight in electric energy, have guaranteed an efficiency of nearly 20%; nevertheless, because of the huge cost, a large-scale development of this technology is still unavailable. It is therefore of great interest to explore whether comparable photovoltaic efficiencies can be achieved using different types of materials that are inexpensive and have the potential for global-scale production. A significant breakthrough was the invention of dye-sensitized solar cells (DSSC) by O’Regan and Grätzel. DSSSC, also known as Grätzel cells, are based on an appropriate photosensitized molecule adsorbed on a semiconductor, generally a metal oxide such as TiO2. The efficiency of such organic DSSC is a topic of paramount importance, from both the fundamental research and especially from application standpoint.
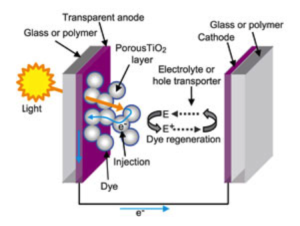
We study the factors that control electron injection, in order to design dyes that provide rapid electron injection and make efficient solar cells.
- AC1921103 Applied Quantum Mechanics and Group theory
- AC1921104 Equilibrium Thermodynamics
- AC1922105 Structural aspects and stereochemistry of inorganic complexes
- AC1922107 Advanced quantum mechanics and molecular modeling
- AN1921103 Quantum Mechanics and Molecular Spectroscopy
- AN1921104 Statistical Mechanics and Molecular Simulations
- AN1922107 Molecular Quantum Mechanics and computational Chemistry
- CH1815108 Physical Chemistry – II
- CH1816111 Physical Chemistry – III
- CH1921103 Quantum Mechanics and Group Theory
- CH1921104 Classical and Statistical Thermodynamics
- CH1922105 Coordination Chemistry
- CH1922107 Chemical Bonding and Computational Chemistry
Chemistry
Department